Publications
* denotes corresponding authors
Refereed Publications from Independent Research at UNC-Chapel Hill
33. Crawford, D.M., Roche, J.C., Guo, Q., Brache, C., & Li, B.* “Pseudomonas Virulence Factor Produces Autoinducer (S)-Valdiazen,” ACS Chemical Biology. (date and pages to be added when available) doi: https://doi.org/10.1021/10.1021/acschembio.4c00837
32. Steiner, O.M., Johnson, R.A., Chen, X., Simke, W.C., & Li, B.* “Activation of Dithiolopyrrolone Antibiotics by Cellular Reductants,” Biochemistry. (2024): 192-202 doi: https://doi.org/10.1021/acs.biochem.4c00533
31. Chen, X., & Li, B.* “Analysis of Co-localized Biosynthetic Gene Clusters Identifies a Membrane-Permeabilizing Natural Product,” Journal of Natural Products (2024): 1694-1703. doi: https://doi.org/10.1021/acs.jnatprod.3c01231
30. Guo, Q., Vitro, C. N., Crawford, D. M., & Li, B.* “A diazeniumdiolate signal in Pseudomonas syringae upregulates virulence factors and promotes survival in plants,” Molecular Plant-Microbe Interactions. (2024): 776–783 doi: https://doi.org/10.1094/MPMI-06-24-0069-R
29. Simke, W.C., Walker, M.E., Calderone, L.C., Putz, A.T., Vitro, C.N., Zizola, C.F., Redinbo, M.R., Pandelia, M-E.*, Grove, T.L.*, & Li, B.* “Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme,” ACS Central Science (2024): 1524-1536 doi: https://doi.org/10.1021/acscentsci.4c00015
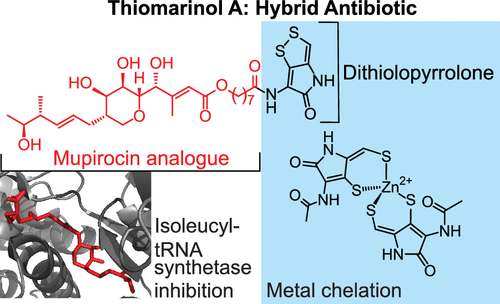
28. Johnson, R.M., Li, K., Chen, X., Morgan, G.L., Aubé, J., & Li, B.* “The Hybrid Antibiotic Thiomarinol A Overcomes Intrinsic Resistance in Escherichia coli Using a Privileged Dithiolopyrrolone Moiety,” ACS Infectious Diseases (2024): 582-593. doi: https://doi.org/10.1021/acsinfecdis.3c00504
27. Chen, X., & Li, B.* “How nature incorporates sulfur and selenium into bioactive natural products,” Current Opinion in Chemical Biology (2023): 102377. doi: https://doi.org/10.1016/j.cbpa.2023.102377
26. Chan, A.N., Chen, X., Falco, J.A., Bak, D.W., Weerapana, E., & Li, B.* “Chemoproteomics Reveals Disruption of Metal Homeostasis and Metalloproteins by the Antibiotic Holomycin,” ACS Chemical Biology (2023): 1909-1914. doi: https://doi.org/10.1021/acschembio.3c00360
25. Guo, Q., Chen, X., & Li, B.* “Purification and characterization of tomato arginine decarboxylase and its inhibition by the bacterial small molecule phevamine A,” Protein Expression and Purification (2023): 106326. doi: https://doi.org/10.1016/j.pep.2023.106326
24. Chen, X., Johnson, R. M., & Li, B.* “A Permissive Amide N-Methyltransferase for Dithiolopyrrolones,” ACS Catalysis 13.3 (2023): 1899-1905. doi: https://doi.org/10.1021/acscatal.2c05439
23. Acken, K. A., & Li, B.* “Pseudomonas virulence factor controls expression of virulence genes in Pseudomonas entomophila,” Plos one 18.5 (2023): e0284907. doi: https://doi.org/10.1371/journal.pone.0284907
22. Lescallette, A. R., Dunn, Z. D., Manning, V. A., Trippe, K. M., & Li, B.* “Biosynthetic origin of formylaminooxyvinylglycine and characterization of the formyltransferase GvgI,” Biochemistry, 61.19 (2022): 2159-2164. doi: https://doi.org/10.1021/acs.biochem.2c00374
21. Patteson, J.B., Fortinez, C.M., Putz, A.T., Rodriguez-Rivas, J., Bryant III, L.H., Adhikari, K., Weigt, M., Schmeing, T.M. & Li, B.* “Structure and Function of a Dehydrating Condensation Domain in Nonribosomal Peptide Biosynthesis,” Journal of the American Chemical Society, 144.31 (2022): 14057-14070. doi: https://doi.org/10.1021/jacs.1c13404
20. Patteson, J. B., Putz, A. T., Tao, L., Simke, W. C., Bryant III, L. H., Britt, R. D., & Li, B.* “Biosynthesis of fluopsin C, a copper-containing antibiotic from Pseudomonas aeruginosa,” Science 374.6570 (2021): 1005-1009. doi: https://www.science.org/doi/full/10.1126/science.abj6749
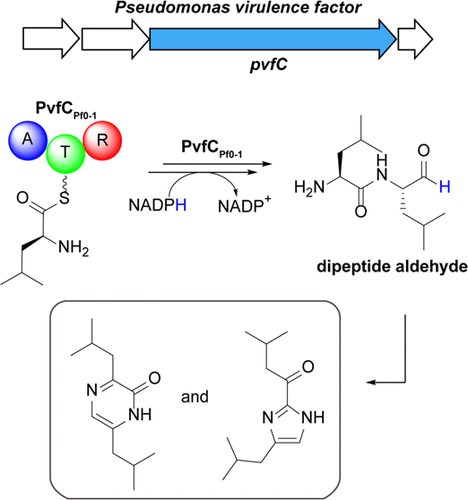
19. Morgan, G. L., Li, K., Crawford, D. M., Aubé, J., & Li, B.* “Enzymatic synthesis of diverse heterocycles by a noncanonical nonribosomal peptide synthetase,” ACS chemical biology 16.12 (2021): 2776-2786. doi: https://doi.org/10.1021/acschembio.1c00623
18. Johnson, R. A., Chan, A. N., Ward, R. D., McGlade, C. A., Hatfield, B. M., Peters, J. M., & Li, B.* “Pseudomonas Virulence Factor Pathway Synthesizes Autoinducers That Regulate the Secretome of a Pathogen,” Journal of the American Chemical Society 143(31), 12003-12013. doi: https://doi.org/10.1021/jacs.1c02622
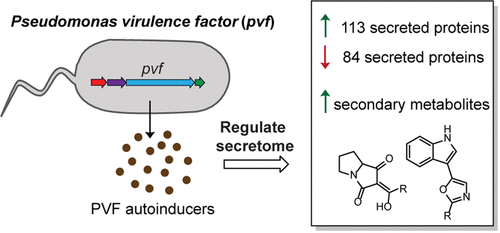
17. Kretsch, A. M., Morgan, G. L., Acken, K. A., Barr, S. A., & Li, B.* “Pseudomonas Virulence Factor Pathway Synthesizes Autoinducers That Regulate the Secretome of a Pathogen,” ACS Chemical Biology 16 (3), 501-509 doi: https://doi.org/10.1021/acschembio.0c00901
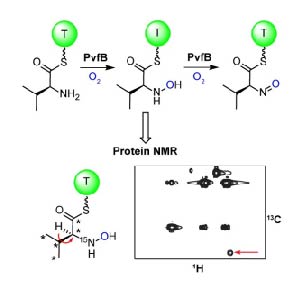
16. Morgan, G. L., Li, B.* “In vitro reconstitution reveals a central role for the N‐oxygenase PvfB in (dihydro)pyrazine‐N‐oxide and valdiazen biosynthesis,” Angew. Chemie. Int. Ed. ASAP doi: https://doi.org/10.1002/ange.202005554
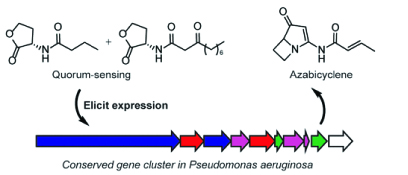
15. Patteson, J. B., Lescallette, A. R., Li, B.* “Discovery and Biosynthesis of Azabicyclene, a Conserved Nonribosomal Peptide in Pseudomonas aeruginosa,” Org. Lett. 13, 4955-4959 doi: https://doi.org/10.1021/acs.orglett.9b01383
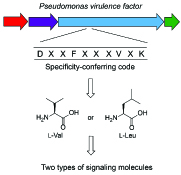
14. Morgan, G. Kretsch, A. M., Santa Maria, K., Weeks, S. J., Li, B.* “Specificity of nonribosomal peptide synthetases in the biosynthesis of the Pseudomonas virulence factor,” Biochemistry ASAP doi: https://doi.org/10.1021/acs.biochem.9b00360
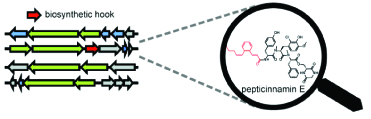
13. Santa Maria, K. C., Chan, A. N., O'Neill, E. M., Li, B.* “Targeted Rediscovery and Biosynthesis of the Farnesyl‐Transferase Inhibitor Pepticinnamin E,” ChemBioChem 20, 1387-1393 (2019) doi: https://doi.org/10.1002/cbic.201900025
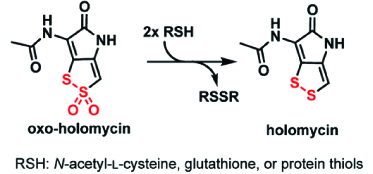
12. Chan, A.N., Wever, W.J., Massolo, E., Allen, S.E., Li, B.* “Reducing holomycin thiosulfonate to its disulfide with thiols,” Chem. Res. Toxicol. (2019), 32, 400-404 doi: 10.1021/acs.chemrestox.8b00243
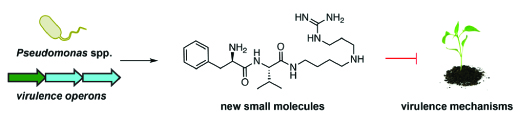
11. O’Neill, E.M., Mucyn, T.S., Patteson, J.B., Finkel, O.M., Chung, E-H., Baccile, J.A., Massolo, E., Schroeder, F.C., Dangl, J.L., Li, B.* “Phevamine A, a bacterial small molecule that suppresses plant immune responses,” Proc. Nat. Acad. Sci. U.S.A., 41, 9514-9522 (2018). doi: 10.1073/pnas.1803779115
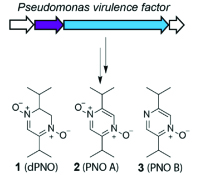
10. Kretsch, A.M., Morgan, G.L., Tyrrell, J., Mevers, E., Vallet-Gély, E., Li, B.* “Discovery of (dihydro)pyrazine N-oxides via genome-mining in Pseudomonas,” Org. Lett., 20, 4791–4795 (2018). DOI: 10.1021/acs.orglett.8b01944
9. Pellock, S., Creekmore, B., Walton, W., Mehta, N., Biernat, K., Cesmat, A., Ariyarathna, Y., Dunn, Z.D., Li, B., Jin, J., James, L., Redinbo, M.* “Piperazine-containing inhibitors intercept the catalytic cycle of gut microbial β-glucuronidases,” ACS Cent. Sci., 4, 868–879 (2018). doi: 10.1021/acscentsci.8b00239
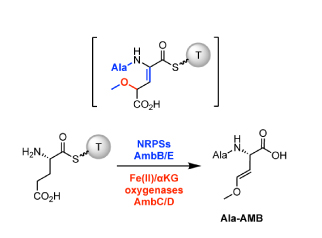
8. Patteson, J.B., Dunn, Z.D., Li, B.* "In Vitro Biosynthesis of the Nonproteinogenic Amino Acid Methoxyvinylglycine," Angew. Chem. Int. Ed. 57, 6780-6785. doi: 10.1002/anie.201713419
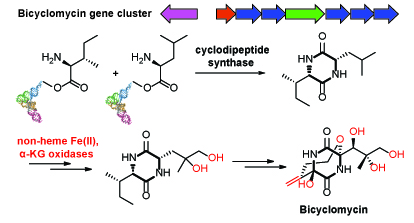
7. Patteson, J.B., Cai, W., Johnson, R.A., Santa Maria, K.C., Li, B.* "Identification of the Biosynthetic Pathway for the Antibiotic Bicyclomycin," Biochemistry, 57, 61–65 (2018). doi: 10.1021/acs.biochem.7b00943
Published in the “Future of Biochemistry” Special Issue
Highlighted in Viewpoint article: Chekan J.R., Moore B.S. “Biosynthesis of the antibiotic bicyclomycin in soil and pathogenic bacteria,” Biochemistry, 57, 897–898 (2018).
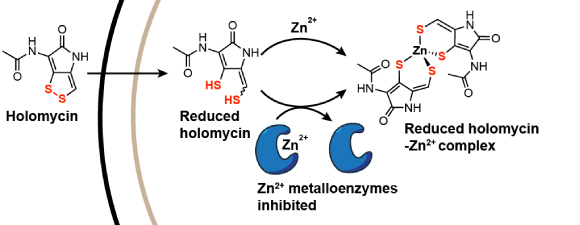
6. Chan, A.N., Shiver, L.S., Wever, W.J., Razvi, S.Z.A., Traxler, M.F., Li, B.* "Role for dithiolopyrrolones in disrupting bacterial metal homeostasis," Proc. Natl. Acad. Sci. U.S.A. 10, 2717-2722 (2017). doi: 10.1073/pnas.1612810114
Highlighted in Chemical and Engineering News: “Mode of action for unusual antibiotic found,” 95, 10 (2017).
5. Shiver, A.L., Osadnik, H., Kritikos, G., Li, B., Krogan, N., Typas, A., Gross, C.A*. “A chemical-genomic screen of neglected antibiotics reveals illicit transport of kasugamycin and blasticidin S”, PLoS Genet., 12, e1006124 (2016). doi: 10.1371/journal.pgen.1006124
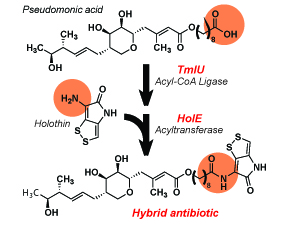
4. Dunn, Z.D., Weaver, W.J., Economou, N.J., Bowers, A.A., Li, B.* "Enzymatic Basis of 'hybridity' in Thiomarinol Biosynthesis," Angew. Chem. Int. Ed. 54, 5137-5141 (2015). doi: 10.1002/anie.201411667
Invited Reviews and commentary
3. Biernat, K.A., Li, B., Redinbo, M.R.* “Microbial unmasking of plant glycosides,” mBio, 9, e02433–17 (2018). (Commentary) doi: 10.1128/mBio.02433-17
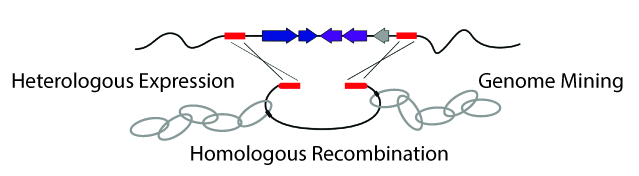
2. Chan, A.N., Santa Maria, K.C., Li, B. "Direct capture technologies for genomics-guided discovery of natural products," Curr. Top. Med. Chem. 15, 1695-1704 (2016). (Review) Link
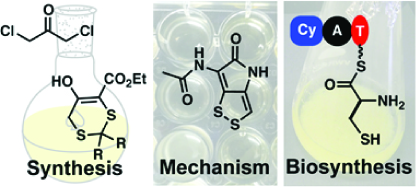
1. Li, B., Weaver, W.J., Walsh, C.T., Bowers, A.A. “Dithiolopyrrolones: Biosynthesis, Synthesis, and Activity of a Unique Class of Disulfide-Containing Antibiotics,” Nat. Prod. Rep. 31, 905-23 (2014) (Review) doi: 10.1039/C3NP70106A
Refereed Publications Prior to UNC-Chapel Hill
9. Ortega, M.A., Cogan, D.P., Mukherjee, S., Garg, N., Li, B., Maffioli, S., Donadio, S., Sosio, M., Escano, J., Smith, J.L., Nair, S.K., and van der Donk, W.A. “Two flavoenzymes install 5-chlorotryptophan and 2-amionvinyl cysteine during the biosynthesis of the lantibiotic NAI-107”, ACS Chem. Bio. 12, 548–557 (2017).
8. Li, B., Forseth R.R., Bowers A.A., Schroeder F.C., Walsh, C.T. "A backup plan for self-protection: S-methylation of holomycin biosynthetic intermediates in Streptomyces clavuligerus." ChemBioChem, 13, 2521-6 (2012). Highlighted in Chem. Eur. J., 18, 15904 (2012).
7. Li, B., Walsh, C.T. "Streptomyces clavuligerus HlmI is an intramolecular disulfide-forming dithiol oxidase in holomycin biosynthesis." Biochemistry, 50, 4615-22 (2011).
6. Li, B., Walsh, C.T. "Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus," Proc. Natl. Acad. Sci. U.S.A., 107, 19731-5 (2010).
5. Li, B., Sher, D., Kelly, L., Shi, Y., Huang, K., Knerr, P.J., Joewono, I., Rusch, D. Chisholm, S.W., van der Donk, W.A. "Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria," Proc. Natl. Acad. Sci. U.S.A., 107, 10430-5 (2010). Highlighted in "A Most Versatile Enzyme," Chemical and Engineering News, 88, 56 (2010). Also listed as the 5th most-read paper online at Proc. Natl. Acad. Sci. U.S.A. in June, 2010
4. Goto, Y., Li, B., Claesen, J., Shi, Y., Bibb, M.J., van der Donk, W.A. "Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights," PLoS Biol., 8, e1000339 (2010).
3. Li, B., Cooper, L.E., van der Donk, W.A. "Chapter 21. In vitro studies of lantibiotic biosynthesis," Methods Enzymol., 458, 533-58 (2009). (Invited Review)
2. Li, B., van der Donk, W.A. "Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin," J. Biol. Chem., 282, 21169-75 (2007).
1. Li, B., Yu, J.P., Brunzelle, J.S., Moll, G.N., van der Donk, W.A., Nair, S.K. "Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis," Science, 311, 1464-7 (2006). Highlighted in perspective "Five golden rings," Science, 311, 1382-3 (2006); "Antimicrobials: A ringing success," Nature Reviews Microbiol., 4, 322-3 (2006); "Resisting the resistance," ACS Chem. Biol., 1, 119 (2006); "Nisin engineered in test tube," Chemical and Engineering News, 84, 9 (2006).
Book Chapter
1. Cooper, L.E., Li, B., van der Donk, W.A. "Biosynthesis and mode of action of lantibiotics," In Comprehensive Natural Products Chemistry II, Eds. Mander, L., Liu, H-w., (May 2010). (Book Chapter)
